Publications
To view a complete list of Dr. Medvedev’s publications, please visit Google Scholar
Selected publications
2025
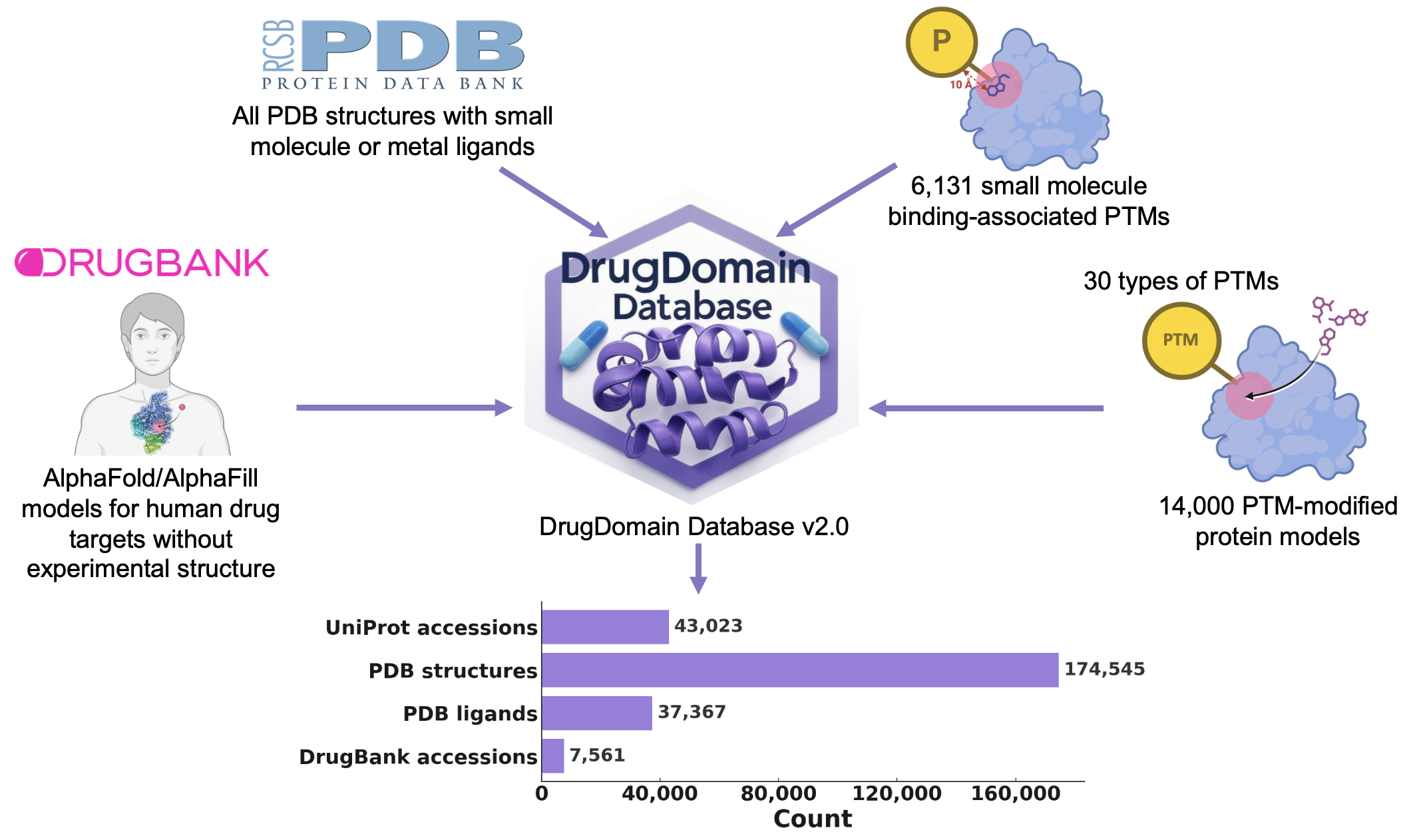
DrugDomain 2.0: comprehensive database of protein domains-ligands/drugs interactions across the whole Protein Data Bank
Computational and Structural Biotechnology Journal
·
01 Sep 2025
·
10.1016/j.csbj.2025.09.018
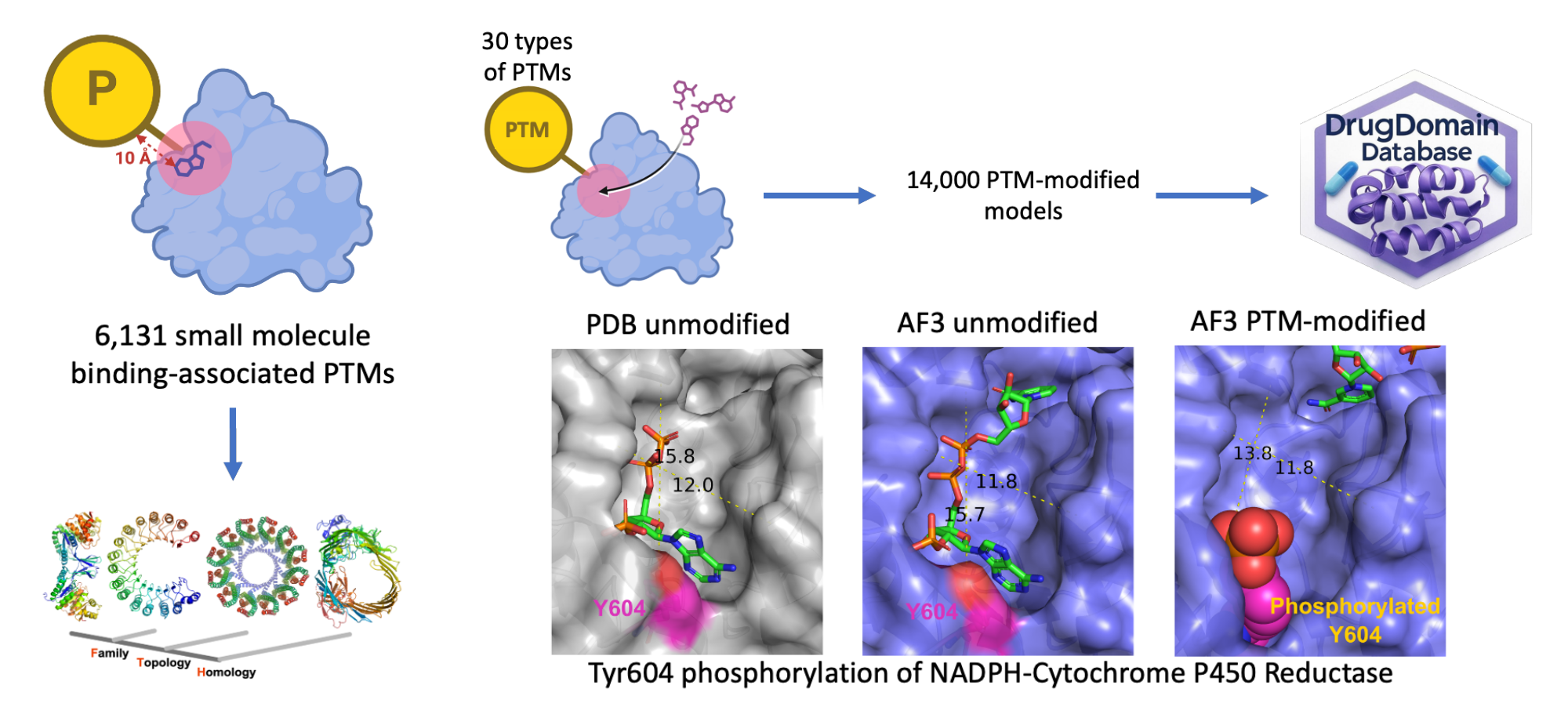
Leveraging AI to explore structural contexts of post-translational modifications in drug binding
Journal of Cheminformatics
·
04 May 2025
·
10.1186/s13321-025-01019-y
2024
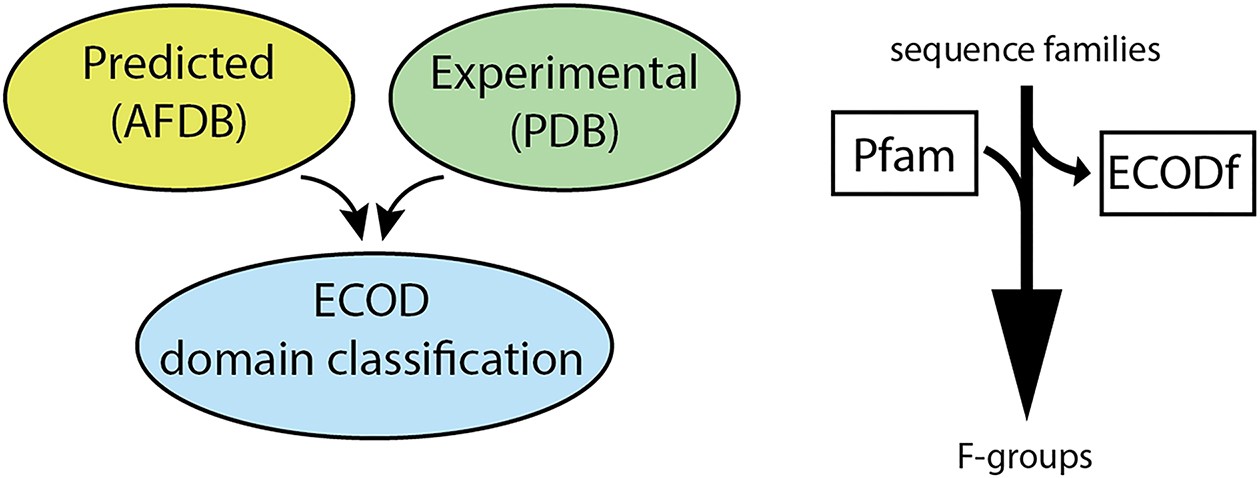
ECOD: integrating classifications of protein domains from experimental and predicted structures
Nucleic Acids Research
·
20 Nov 2024
·
doi:10.1093/nar/gkae1029

Protein Science
·
09 Jul 2024
·
doi:10.1002/pro.5116
Structure classification of the proteins from Salmonella enterica pangenome revealed novel potential pathogenicity islands
Scientific Reports
·
28 May 2024
·
doi:10.1038/s41598-024-60991-x
ECOD domain classification of 48 whole proteomes from AlphaFold Structure Database using DPAM2
PLOS Computational Biology
·
28 Feb 2024
·
doi:10.1371/journal.pcbi.1011586
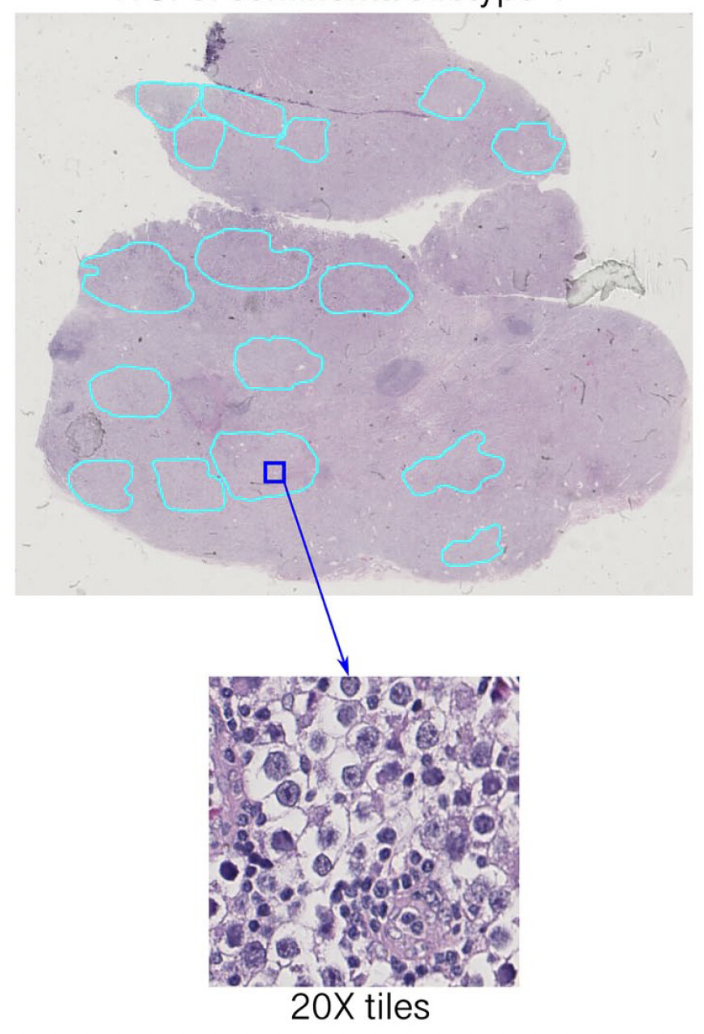
Deep Learning for Subtypes Identification of Pure Seminoma of the Testis
Clinical Pathology
·
01 Jan 2024
·
doi:10.1177/2632010X241232302
2023
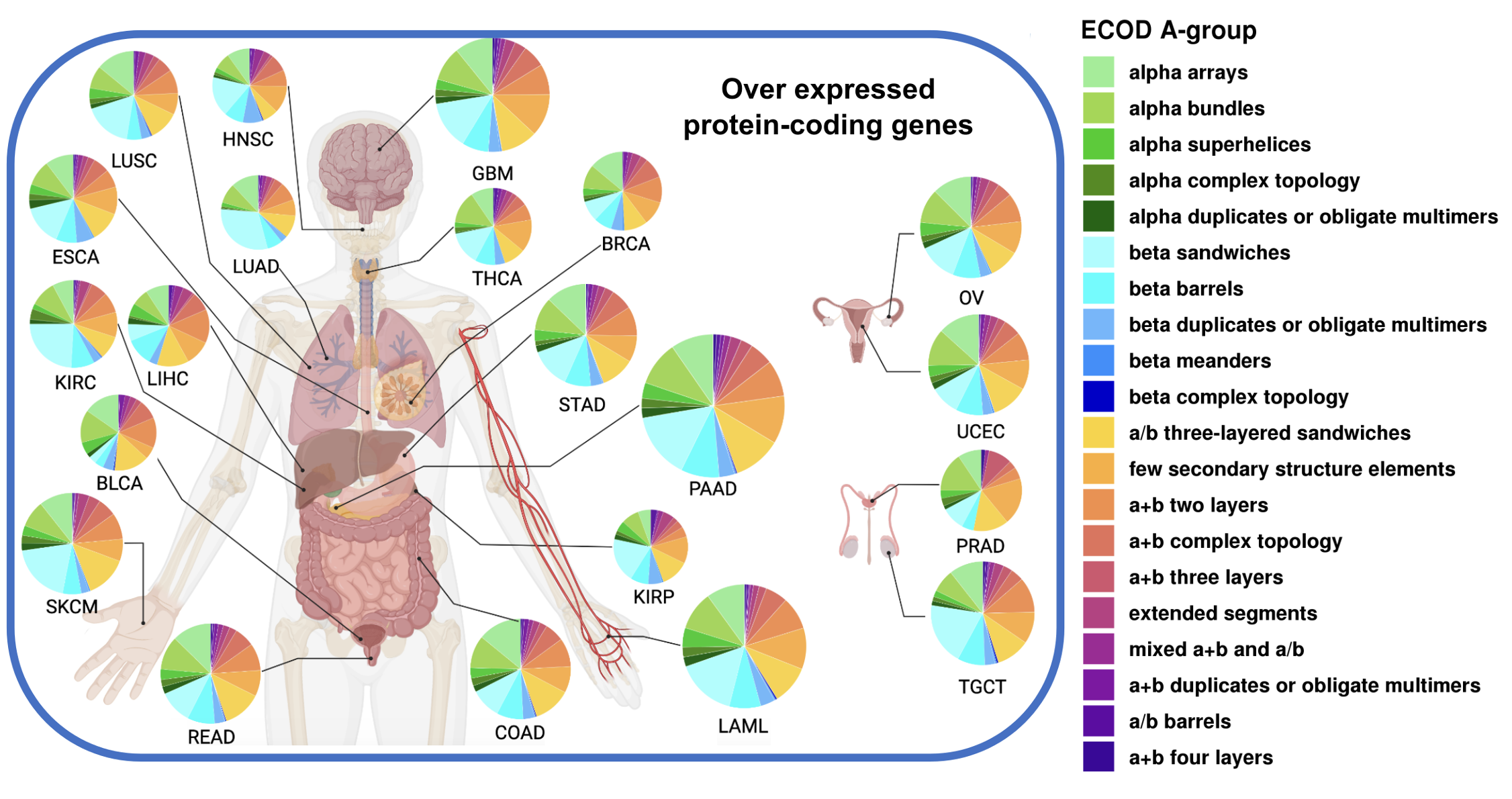
Pan-cancer structurome reveals overrepresentation of beta sandwiches and underrepresentation of alpha helical domains
Scientific Reports
·
25 Jul 2023
·
doi:10.1038/s41598-023-39273-5
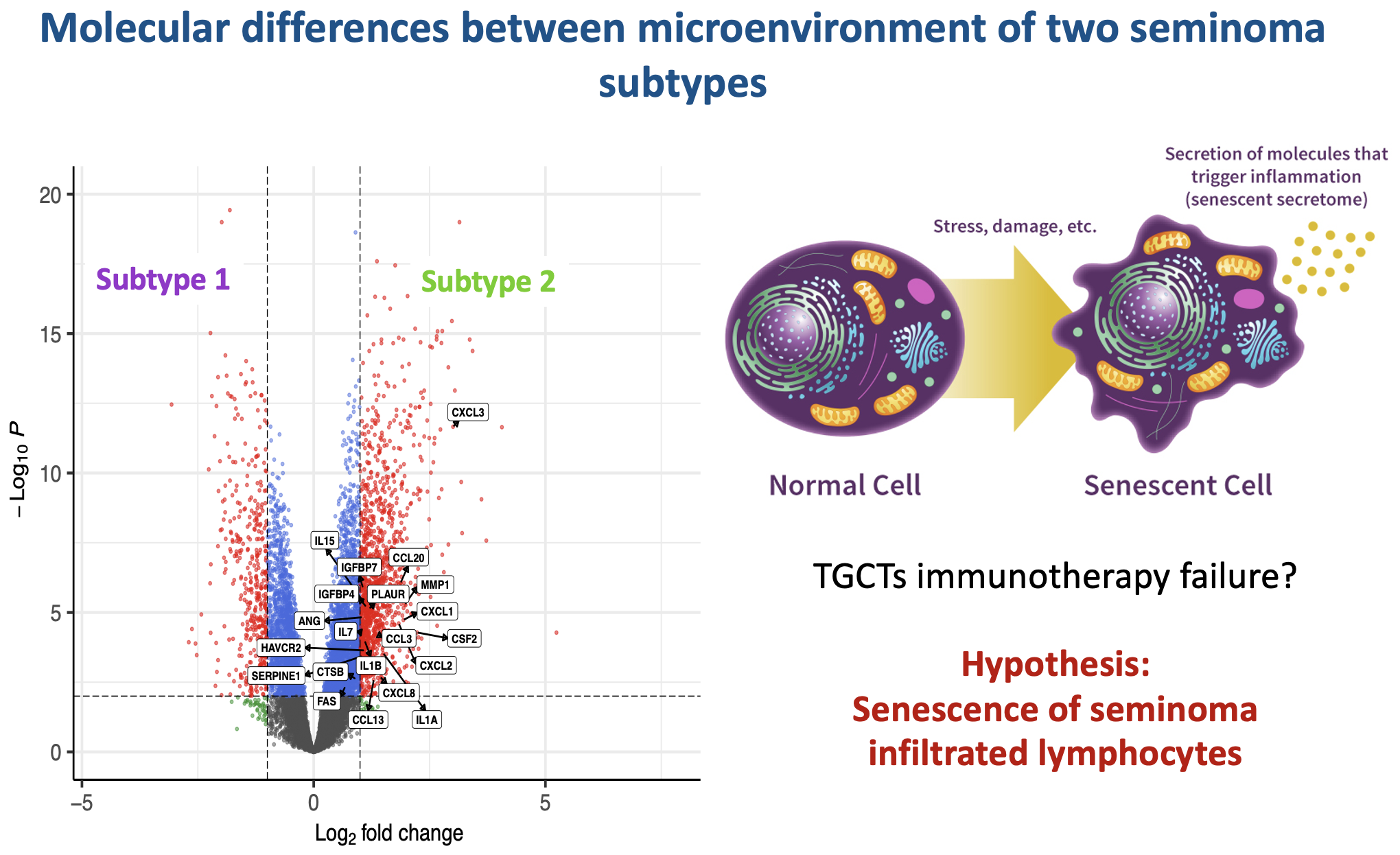
Seminoma subtypes differ in the organization and functional state of the immune microenvironment
3 Biotech
·
01 Mar 2023
·
doi:10.1007/s13205-023-03530-1
2022
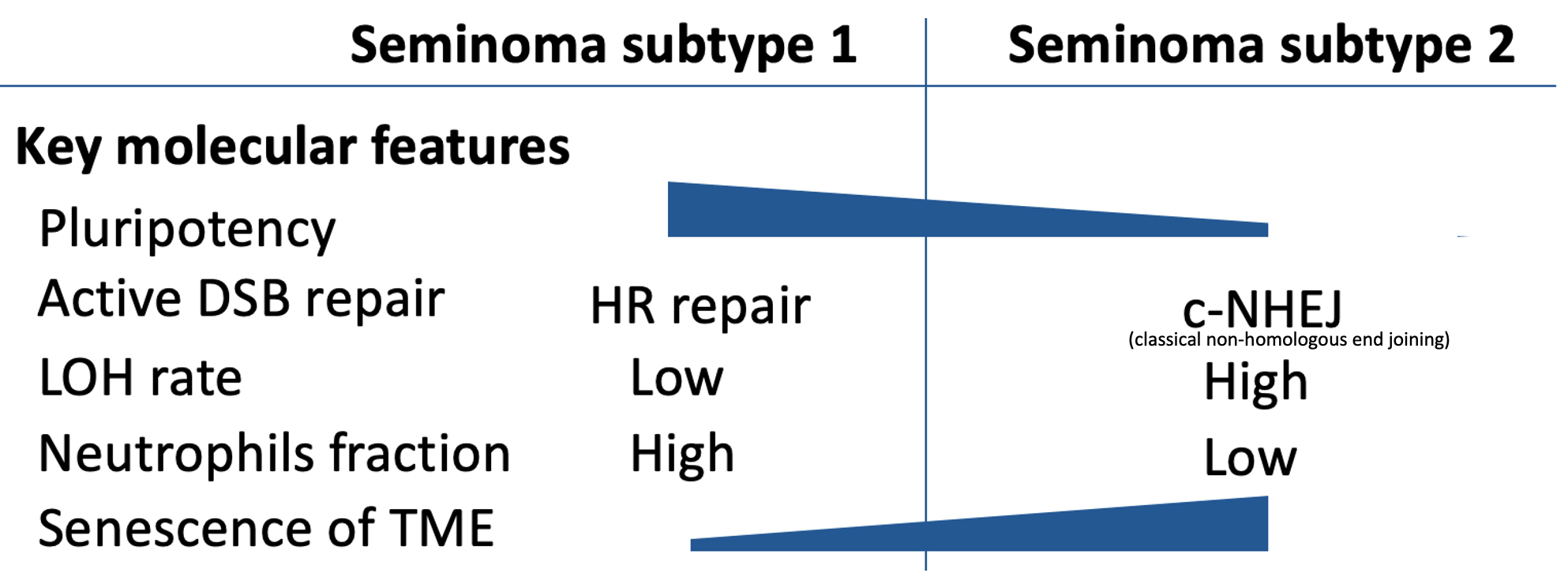
Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis
Cancer Informatics
·
01 Jan 2022
·
doi:10.1177/11769351221132634
2021
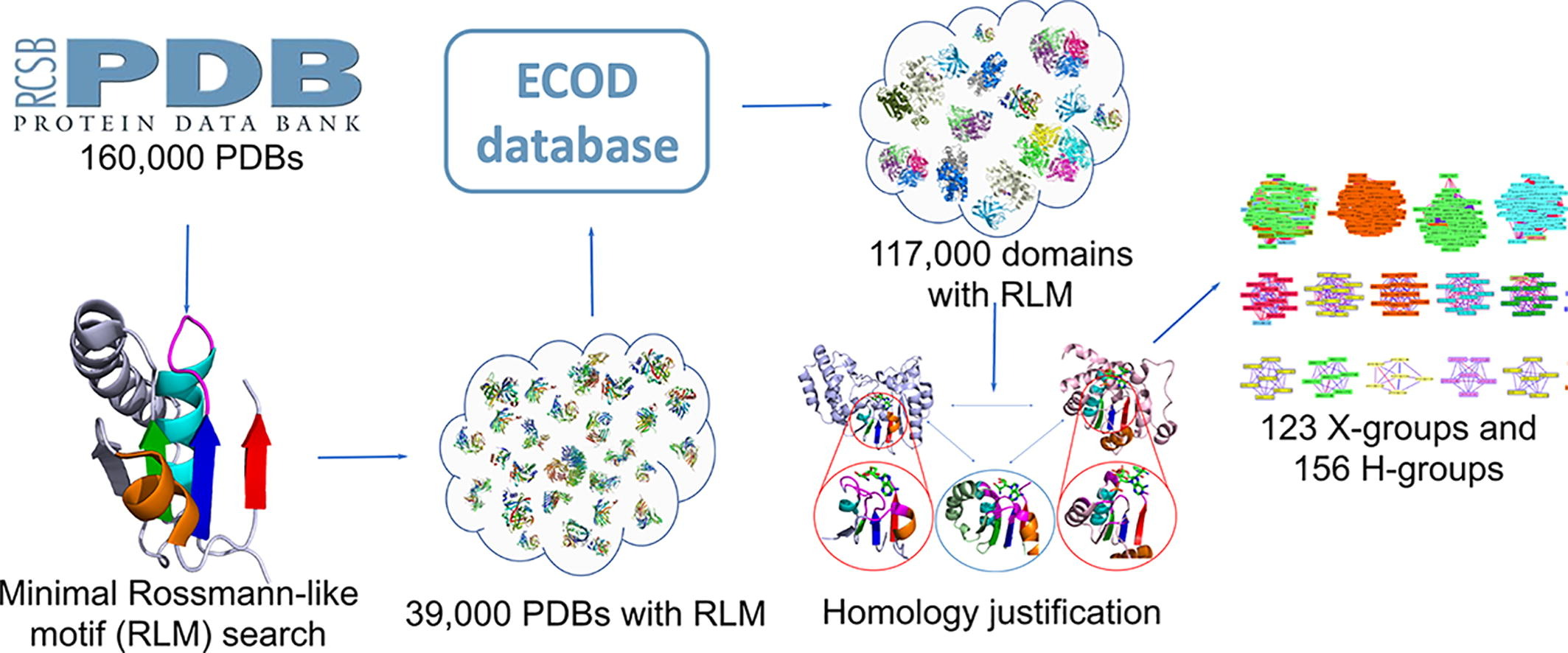
A Fifth of the Protein World: Rossmann-like Proteins as an Evolutionarily Successful Structural unit
Journal of Molecular Biology
·
01 Feb 2021
·
doi:10.1016/j.jmb.2020.166788
2019
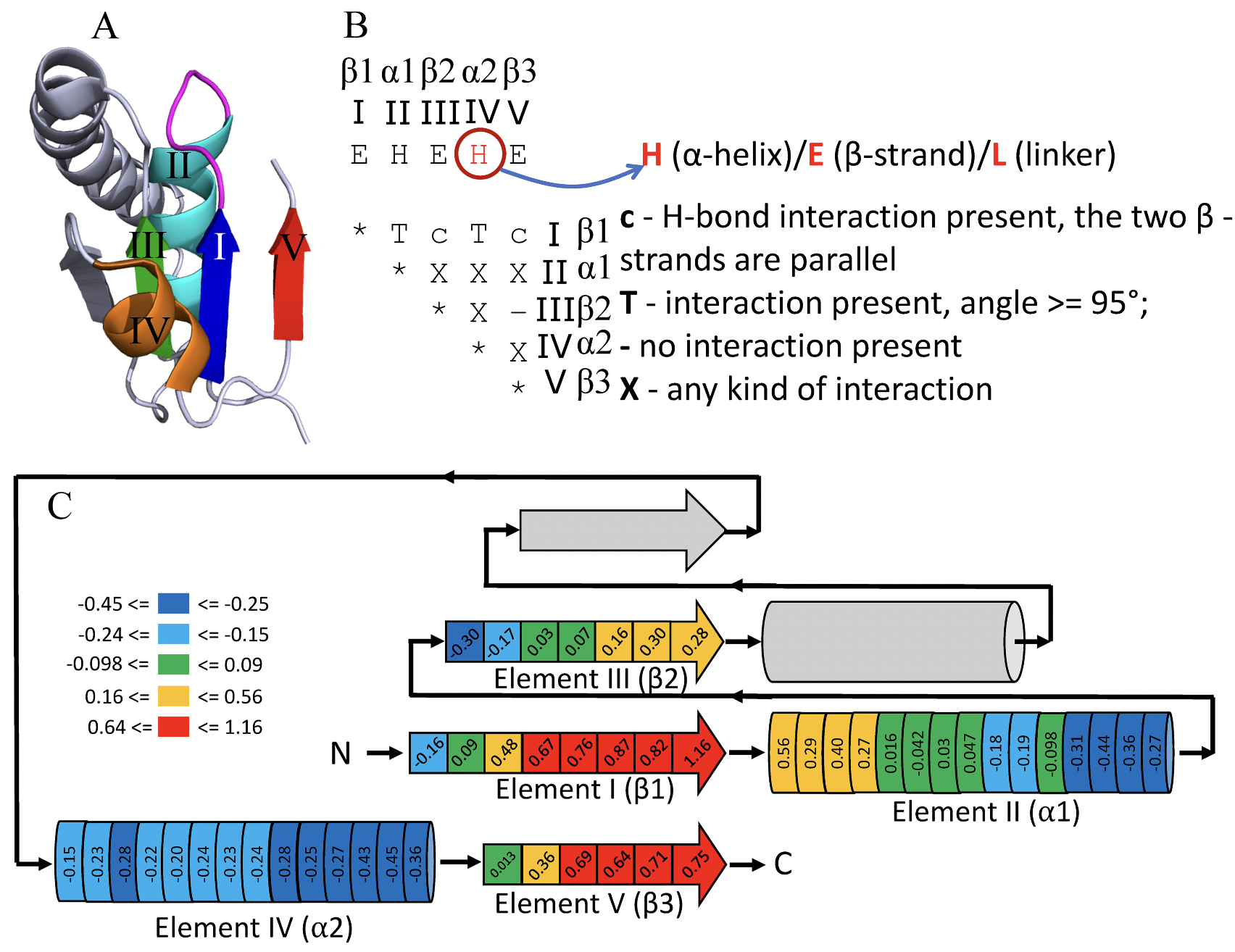
Functional analysis of Rossmann-like domains reveals convergent evolution of topology and reaction pathways
PLOS Computational Biology
·
23 Dec 2019
·
doi:10.1371/journal.pcbi.1007569