Our research
Our research employs a multiscale approach to develop and apply methods for the classification and analysis of large-scale biomedical datasets.
- At the molecular level, we focus on developing tools and databases to classify and analyze protein domain features and their interactions with small molecules and drugs.
- At the cellular level, we focus on analyzing expression profile variations within cancer types to uncover the factors that lead to diverse clinical outcomes.
- At the tissue level we develop deep learning methods to identify cancer subtypes using tissue histopathology and linking tissue morphology to molecular features to improve diagnosis and treatment selection.
Structural Contexts of Post-Translational Modifications in Drug Binding
Understanding the role of post-translational modifications in modulating protein function and drug interactions is crucial for advancing drug discovery. To address this, I have developed a first-of-its-kind resource leveraging recent advances in AI-powered protein structure prediction to map thousands of modifications to their drug targets across the human proteome. Our initial investigations reveal significant effects, such as the structural destabilization of a drug-binding site by a cancer-related phosphorylation. All data and generated models are available from DrugDomain database.
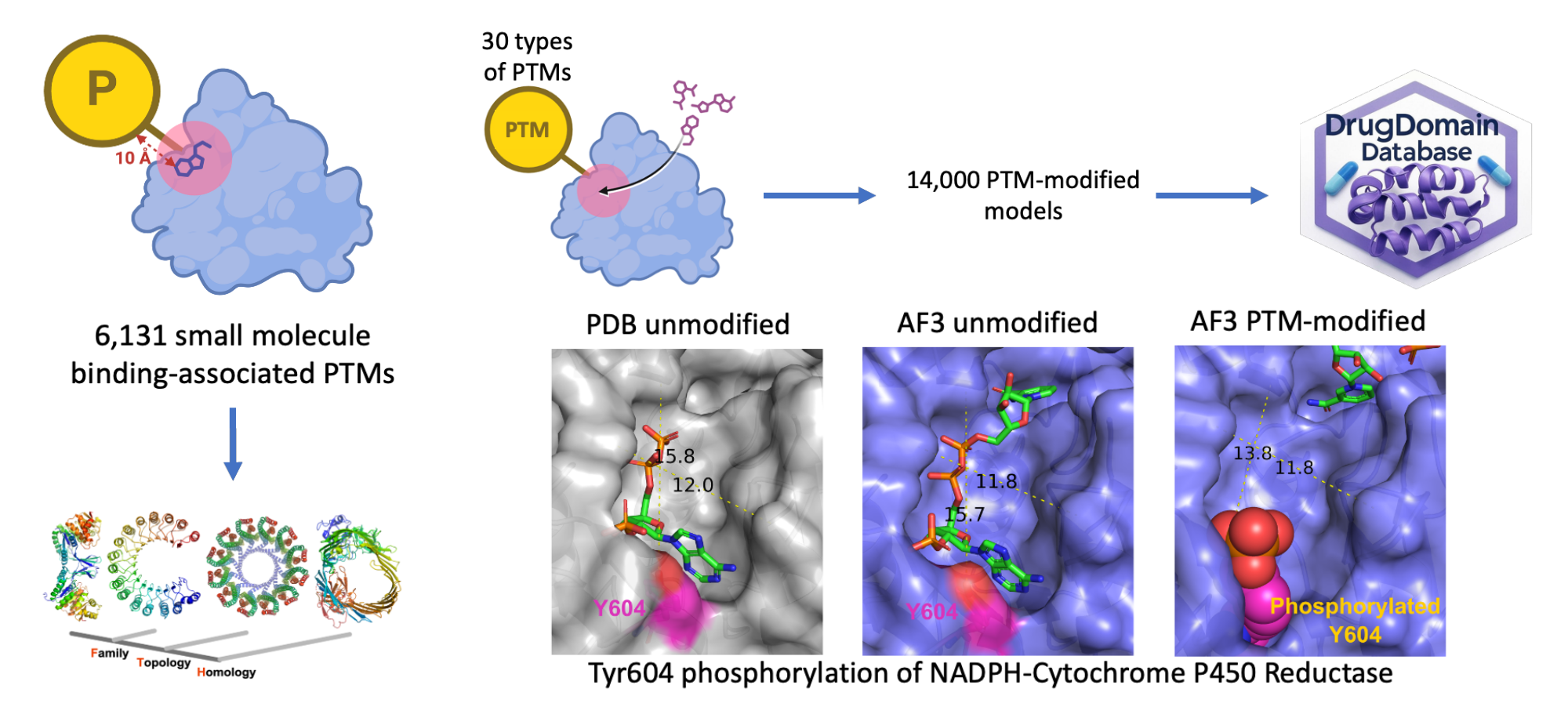
DrugDomain database and drug discovery
DrugDomain database reports domains of proteins that are targets for small molecules and drugs. It includes both experimentally determined PDB structures and AlphaFold models enriched with ligands from experimental data. We plan to use DrugDomain database for identification of novel protein targets for drug repurposing and off-target interaction effects based on evolutionary classification of protein domains.

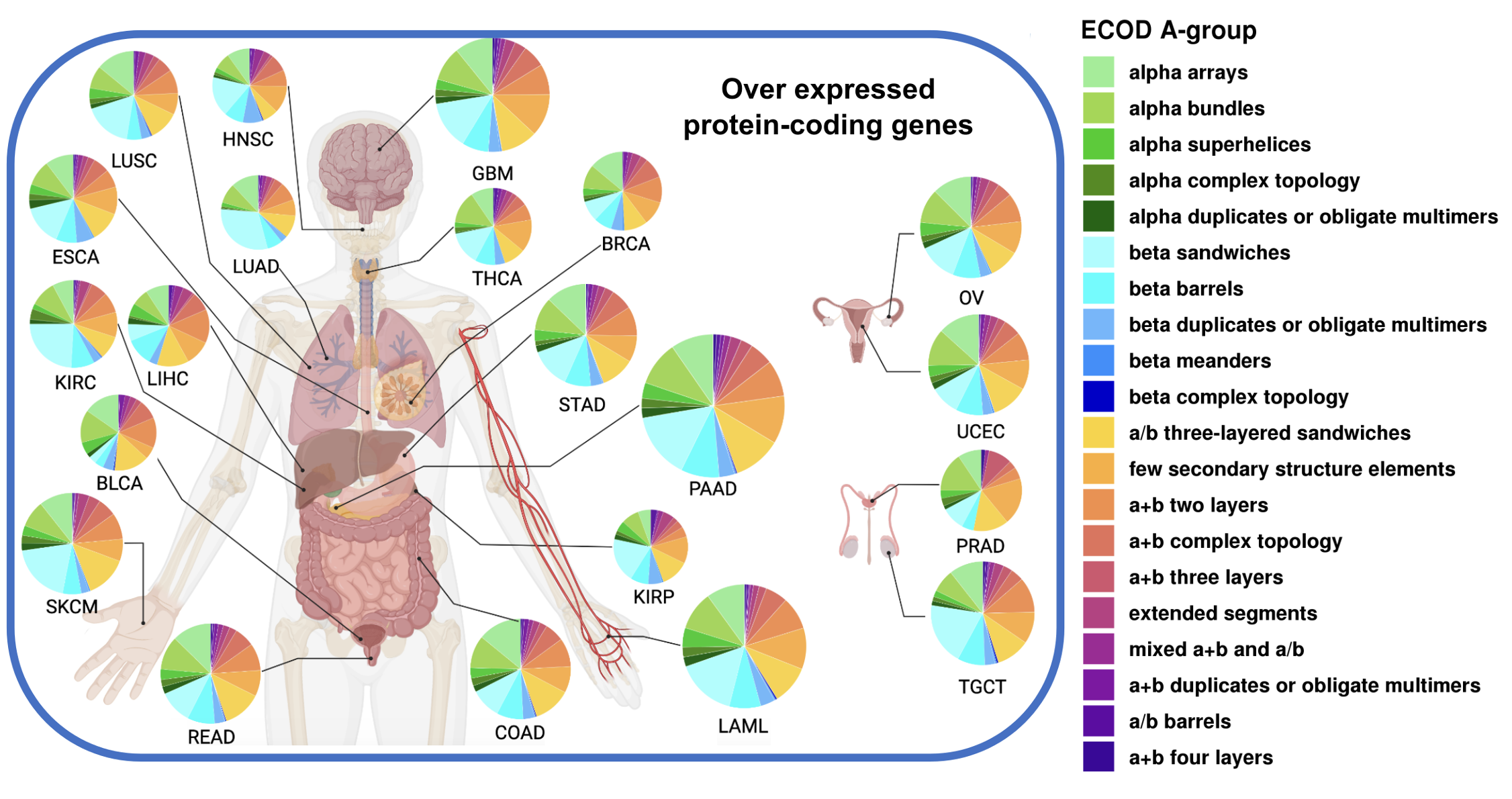
Pan-cancer structurome
The revolutionary accuracy of AlphaFold has dramatically expanded our knowledge of the pan-cancer structurome. By examining the domain architectures of proteins dysregulated in 21 cancer types, we’ve uncovered a consistent pattern: an increase in beta sandwich domains, possibly fueled by tumor-related inflammation, and a decrease in alpha helical domains essential for cellular equilibrium. These findings demonstrate how cancer hijacks the protein structural landscape, providing new insights into disease mechanisms and potential therapeutic targets.
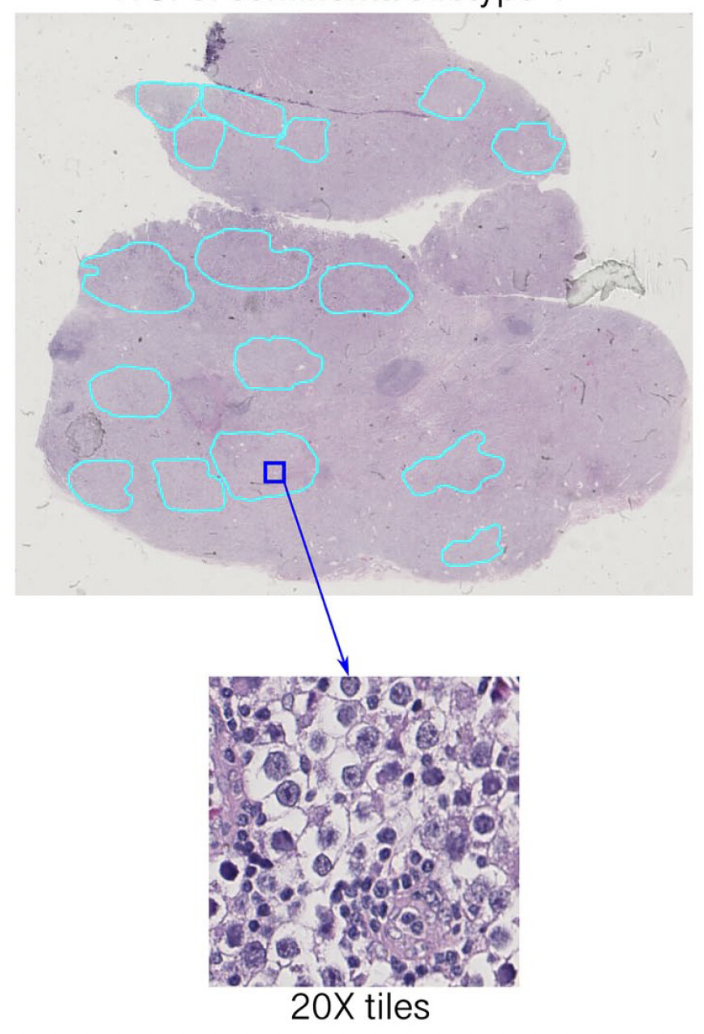
Identification of cancer subtypes
Testicular germ cell tumors, a leading cancer in young men, present a treatment challenge with heterogeneous responses to platinum therapy. By dissecting the molecular landscape of seminomas, the most common TGCT subtype, I’ve identified two distinct subtypes with differing developmental states and DNA repair mechanisms. Notably, our analysis of the tumor microenvironment reveals that one subtype shows signs of immune cell senescence, offering another potential explanation for immunotherapy resistance and highlighting the need for subtype-specific treatment strategies.
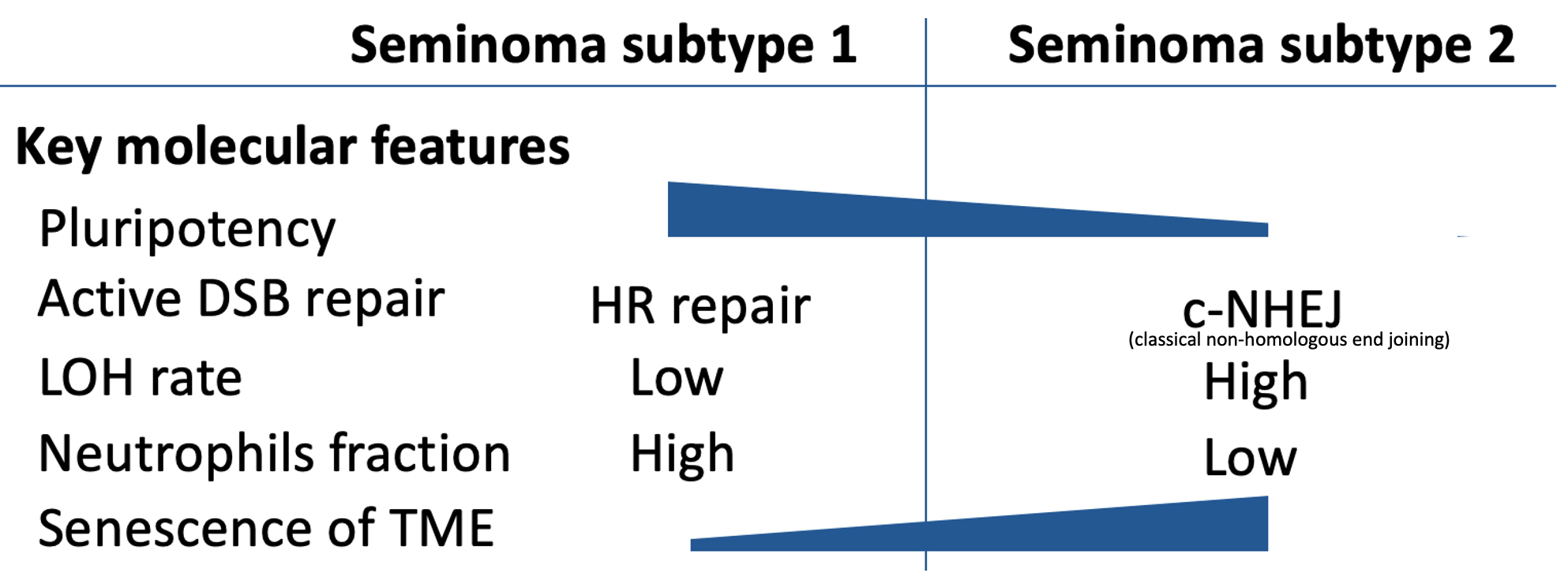
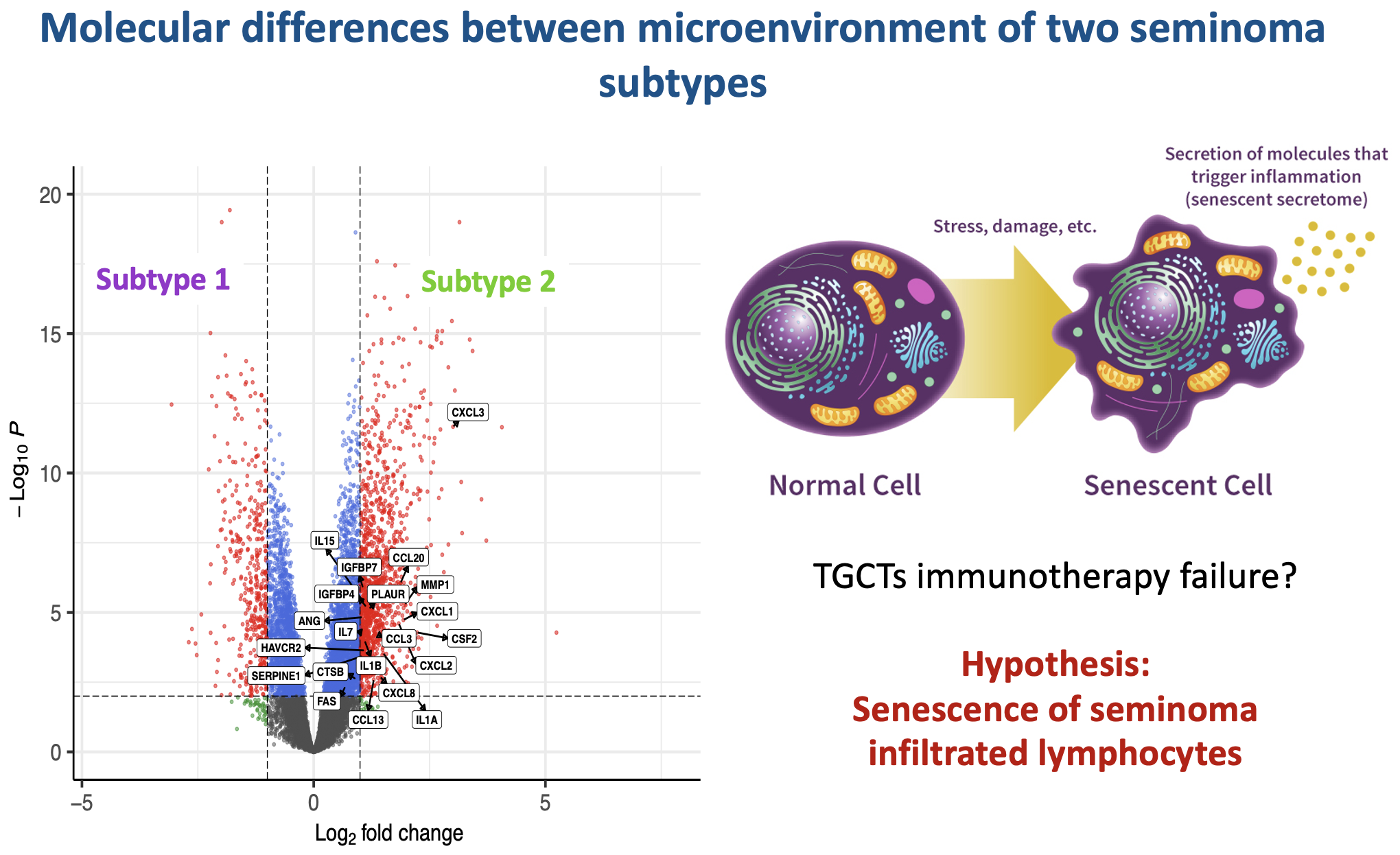
Digital Pathology Because clinical pathologists failed to distinguish seminoma subtypes on immunohistochemical slides, I have developed a deep learning (DL) image classifier for the identification of seminoma subtypes using histopathological slides. The results revealed histopathological differences between the two subtypes of pure seminoma. These findings provide additional confirmation and support the notion that seminoma can be further stratified into distinct subtypes.